
Breeders have always used pedigrees to manage the genetics of Thoroughbred horses. Pedigree analysis is firmly rooted in our understanding of classical, Mendelian genetics. However, pedigrees only track relationships. The actual genetic variation present in any animal can only be assessed by reading the DNA itself.
In 2006, the whole genome sequence of the horse was first reported. It was an expensive undertaking, but since then the costs of doing this work decreased dramatically such that sequencing the whole genome of a horse is a routine activity in research laboratories.
The DNA sequence contains information about the extent of genetic variation, relationship to other horses, relationship to other breeds, levels of inbreeding and can even provide raw material for discovery to potentially deleterious genes that interfere with the success of the breeder.
On August 6, 2021, Teruaki Tozaki and colleagues from the Laboratory of Racing Chemistry and Japan Racing Association published a landmark paper describing whole genome sequencing of 101 Thoroughbreds in Japan. While scientists have been identifying DNA variation in short regions for the last three decades, this study is unique in that these scientists collected data on all 2.41 billion DNA nucleotides of the 101 horses.
This work provides a baseline for comparison of different populations of Thoroughbreds as well as a benchmark to assess changes over time. In addition, we can compare the variation found in this study to variation found in other breeds. Jagannathan and colleagues (2019 Anim. Genet. 50, 74–77) sequenced the whole genomes of 88 horses of diverse breeds including Warmblood, Standardbred, Quarter Horse, Arabian, Morgan, Franch-es-Montagne, Paint, Icelandic, Shetland, Akhal-Teke, Noriker, Welsh ponies and one Thoroughbred. The two studies were similar in design, so we can directly compare their results such as those shown in Table 1.
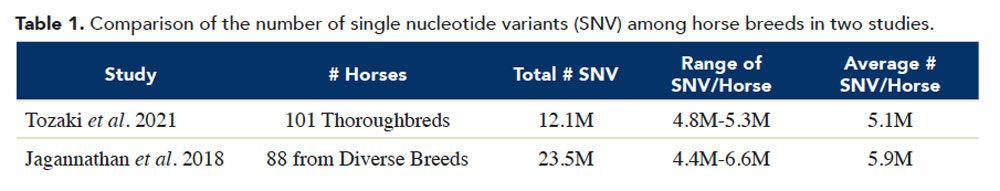
The total numbers of single nucleotide variants (SNVs) found in the Jagannathan study was almost twice as large as those found in the Tozaki study. This illustrates the great amount of diversity existing among horses of all breeds. However, when we examine the number of SNVs found in each horse (Max-Min), Thoroughbred horses fall within the range for the diversity of breeds. Specifically, while the Jagannathan study reported a range of 4.4 million-6.6 million SNV per animal, the Thoroughbred counts fall within that range, 4.8 million-5.3 million.
Two technical caveats bear mentioning here. SNVs are just one type of DNA variant. Other types of variants exist, including DNA insertions, deletions and repeats. Therefore, the total number of variants including those in other categories is certainly greater than the number of SNVs reported.
Another—and perhaps very consequential—caveat is that the number of SNVs were determined through comparisons with reference to the genome of a Thoroughbred mare (the equine reference genome). If we were to use a different breed as a reference—say a Shire horse—we will see a larger number of variants for Thoroughbred horses when compared to this new reference, but less for Shires.
Regardless of how we count the variants, these results suggest that the amount of variation found among Thoroughbreds is not exceptionally depleted when compared to the range of variation among other horse breeds.
Arguably, some of the most important outcomes of this study are yet-to-be generated products of these data. For instance:
- The information serves as a baseline of diversity to assess and model/predict changes in the future population resulting from current and evolving breeding practices.
- The 12.1 million genetic variants identified among these 101 Thoroughbreds can be assessed to determine which might cause fitness problems, and those desirable for health and racing performance.
- These data can be applied to assist in detecting inappropriate modifications of DNA, called gene doping, done in order to enhance racing performance.
You can contact the researchers here:
- Ernest Bailey, PhD, ebailey@uky.edu, 859-218-1105; Maxwell H. Gluck Equine Research Center University of Kentucky, Lexington, Kentucky
- Ted Kalbfleisch, PhD, ted.kalbfleisch@uky.edu, 859-218-1147 Maxwell H. Gluck Equine Research Center University of Kentucky, Lexington, Kentucky
- Jessica Petersen, PhD, Jessica.petersen@unl.edu, 402-472-6328 Department of Animal Science, University of Nebraska-Lincoln, Lincoln, Nebraska.
This report was first published in the January 2022 Equine Disease Quarterly from the Gluck Equine Research Center at the University of Kentucky.








